Chapter 7 Data Wrangling
In data analysis, having known and expected patterns in the data is important. Wrangling is the act of getting messy real world data organized for analysis.
Figure 7.1: Visualizing messy data.
 |
At the end of this chapter you should be able to
|
7.1 Tidy Data
Tidy data is a concept in data science and statistics that refers to a specific way of organizing data into a tabular format that is easy to work with. The concept of tidy data was introduced by Hadley Wickham in his paper “Tidy Data” published in the Journal of Statistical Software in 2014.
In tidy data, each row represents a single observation or record, and each column represents a single variable or attribute. Additionally, there should be no duplicate columns or rows, and each cell should contain a single value.
More specifically, tidy data should have the following properties:
Each variable has its own column: In tidy data, each variable or attribute should have its own column. This makes it easy to compare and analyze different variables.
Each observation has its own row: In tidy data, each observation or record should have its own row. This makes it easy to perform calculations on individual observations or groups of observations.
There should be no duplicate columns or rows: In tidy data, there should be no duplicate columns or rows. This ensures that the data is organized in a clear and concise manner.
Each cell should contain a single value: In tidy data, each cell should contain a single value. This makes it easy to perform calculations and manipulate the data.
By organizing data in a tidy format, it becomes easier to analyze and visualize the data using tools like R and Python. Additionally, tidy data is more robust and less prone to errors than other types of data formats, making it easier to work with and share with others.
 |
1.2.1 Online Cheat-sheets |
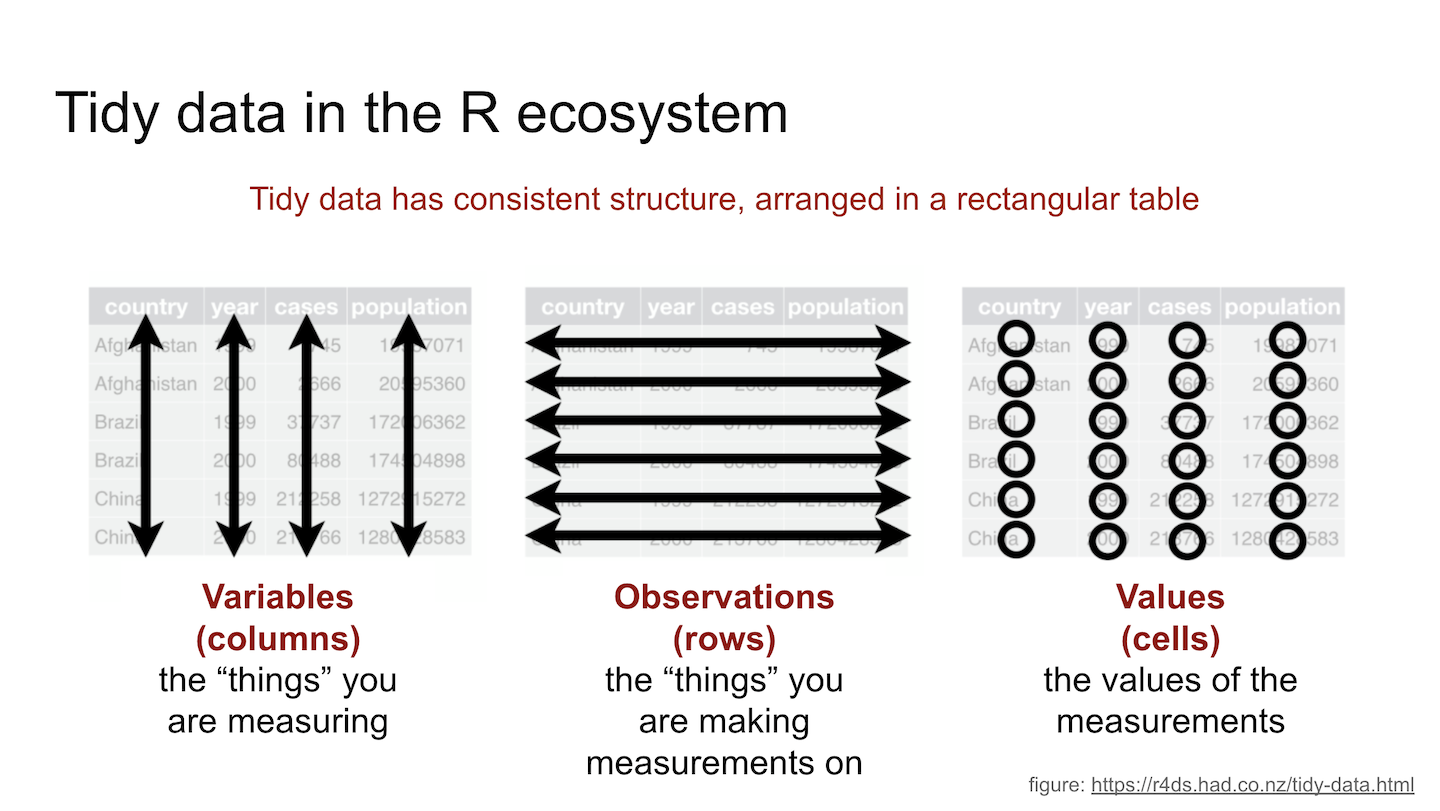
Figure 7.2: Visualizing tidy data concepts.
7.2 Example Walkthrough
We have a processed MS data set of metabolites measured for different bacteria at different time-points with different dosages of an antibiotic.
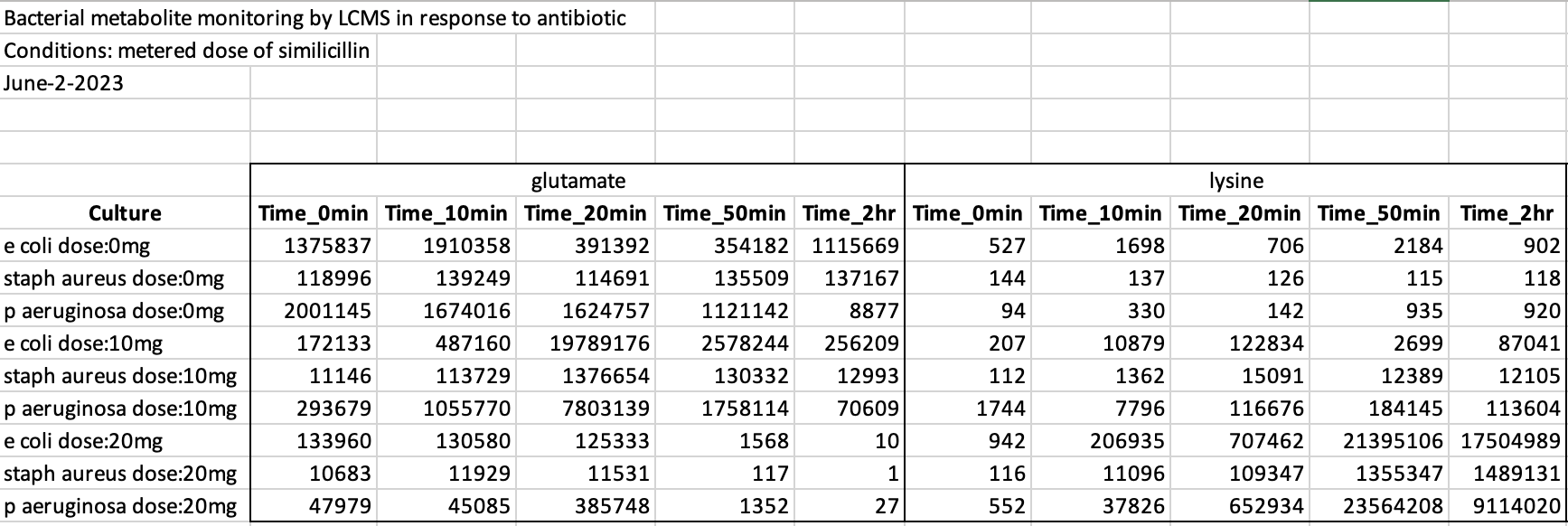
Figure 7.3: Checking our messy data table.
Having a look at the data, we can see definitely that it is not tidy.
Our goal will be to:
* Read the data (in wide, untidy format)
* In a single dplyr pipe create a tidy with culture, dose_mg, metabolite, time_min, abundance as our columns.
read_
Download the data …
url <- "https://raw.githubusercontent.com/jeffsocal/ASMS_R_Basics/main/data/bacterial-metabolites_dose-simicillin_wide.csv"
download.file(url, destfile = "./data/bacterial-metabolites_dose-simicillin_wide.csv")Let’s start with the read-in.
library(tidyverse)
tbl_met <- read_csv("data/bacterial-metabolites_dose-simicillin_wide.csv")
tbl_met## # A tibble: 9 × 21
## Culture glutamate_Time_0min glutamate_Time_10min glutamate_Time_20min
## <chr> <dbl> <dbl> <dbl>
## 1 e coli dose:0… 191666 2082823 1796910
## 2 staph aureus … 125210 132156 114445
## 3 p aeruginosa … 978126 397012 267599
## 4 e coli dose:1… 1445 1017467 14456533
## 5 staph aureus … 152 128683 1360154
## 6 p aeruginosa … 23 1612622 18282386
## 7 e coli dose:2… 885 42356 11758951
## 8 staph aureus … 136 4276 1162855
## 9 p aeruginosa … 3098 68146 19333030
## # ℹ 17 more variables: glutamate_Time_50min <dbl>, glutamate_Time_2hr <dbl>,
## # lysine_Time_0min <dbl>, lysine_Time_10min <dbl>, lysine_Time_20min <dbl>,
## # lysine_Time_50min <dbl>, lysine_Time_2hr <dbl>,
## # `succinic acid_Time_0min` <dbl>, `succinic acid_Time_10min` <dbl>,
## # `succinic acid_Time_20min` <dbl>, `succinic acid_Time_50min` <dbl>,
## # `succinic acid_Time_2hr` <dbl>, phosphatidylcholine_Time_0min <dbl>,
## # phosphatidylcholine_Time_10min <dbl>, …separate
Let’s separate Culture into two new variables: Species and Dose.
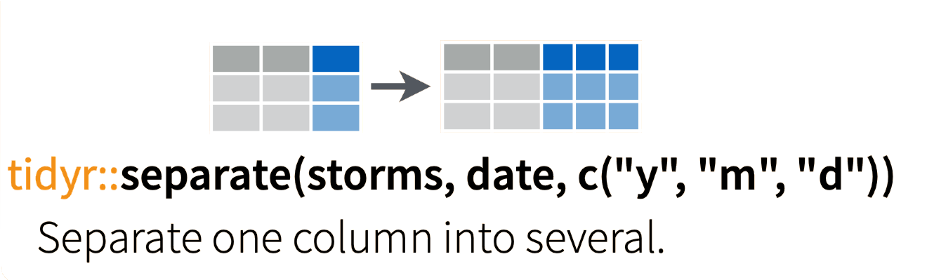
Figure 7.4: Separating columns in a tibble.
## # A tibble: 9 × 22
## culture dose_mg glutamate_Time_0min glutamate_Time_10min
## <chr> <chr> <dbl> <dbl>
## 1 e coli 0mg 191666 2082823
## 2 staph aureus 0mg 125210 132156
## 3 p aeruginosa 0mg 978126 397012
## 4 e coli 10mg 1445 1017467
## 5 staph aureus 10mg 152 128683
## 6 p aeruginosa 10mg 23 1612622
## 7 e coli 20mg 885 42356
## 8 staph aureus 20mg 136 4276
## 9 p aeruginosa 20mg 3098 68146
## # ℹ 18 more variables: glutamate_Time_20min <dbl>,
## # glutamate_Time_50min <dbl>, glutamate_Time_2hr <dbl>,
## # lysine_Time_0min <dbl>, lysine_Time_10min <dbl>, lysine_Time_20min <dbl>,
## # lysine_Time_50min <dbl>, lysine_Time_2hr <dbl>,
## # `succinic acid_Time_0min` <dbl>, `succinic acid_Time_10min` <dbl>,
## # `succinic acid_Time_20min` <dbl>, `succinic acid_Time_50min` <dbl>,
## # `succinic acid_Time_2hr` <dbl>, phosphatidylcholine_Time_0min <dbl>, …mutate
Still some issues here. We managed a separation but out dose column isn’t numeric! We need to remove the “mg”.
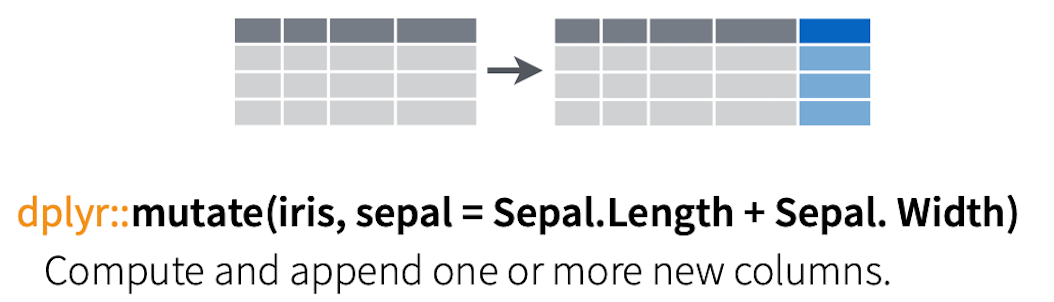
Figure 7.5: Mutate:Making new columns or changing existing ones.
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(Dose = str_remove(dose_mg, "mg"))## # A tibble: 9 × 23
## culture dose_mg glutamate_Time_0min glutamate_Time_10min
## <chr> <chr> <dbl> <dbl>
## 1 e coli 0mg 191666 2082823
## 2 staph aureus 0mg 125210 132156
## 3 p aeruginosa 0mg 978126 397012
## 4 e coli 10mg 1445 1017467
## 5 staph aureus 10mg 152 128683
## 6 p aeruginosa 10mg 23 1612622
## 7 e coli 20mg 885 42356
## 8 staph aureus 20mg 136 4276
## 9 p aeruginosa 20mg 3098 68146
## # ℹ 19 more variables: glutamate_Time_20min <dbl>,
## # glutamate_Time_50min <dbl>, glutamate_Time_2hr <dbl>,
## # lysine_Time_0min <dbl>, lysine_Time_10min <dbl>, lysine_Time_20min <dbl>,
## # lysine_Time_50min <dbl>, lysine_Time_2hr <dbl>,
## # `succinic acid_Time_0min` <dbl>, `succinic acid_Time_10min` <dbl>,
## # `succinic acid_Time_20min` <dbl>, `succinic acid_Time_50min` <dbl>,
## # `succinic acid_Time_2hr` <dbl>, phosphatidylcholine_Time_0min <dbl>, …Ok, the “mg” is removed but if we check the tibble, Dose is still chr, so it’s still computed as text by R! Let’s make it numeric.
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg))## # A tibble: 9 × 22
## culture dose_mg glutamate_Time_0min glutamate_Time_10min
## <chr> <dbl> <dbl> <dbl>
## 1 e coli 0 191666 2082823
## 2 staph aureus 0 125210 132156
## 3 p aeruginosa 0 978126 397012
## 4 e coli 10 1445 1017467
## 5 staph aureus 10 152 128683
## 6 p aeruginosa 10 23 1612622
## 7 e coli 20 885 42356
## 8 staph aureus 20 136 4276
## 9 p aeruginosa 20 3098 68146
## # ℹ 18 more variables: glutamate_Time_20min <dbl>,
## # glutamate_Time_50min <dbl>, glutamate_Time_2hr <dbl>,
## # lysine_Time_0min <dbl>, lysine_Time_10min <dbl>, lysine_Time_20min <dbl>,
## # lysine_Time_50min <dbl>, lysine_Time_2hr <dbl>,
## # `succinic acid_Time_0min` <dbl>, `succinic acid_Time_10min` <dbl>,
## # `succinic acid_Time_20min` <dbl>, `succinic acid_Time_50min` <dbl>,
## # `succinic acid_Time_2hr` <dbl>, phosphatidylcholine_Time_0min <dbl>, …pivot_
Now, we have a bigger problem… multiple pieces of information encoded into each column!
We need this tibble to be long rather than wide to by tidy. We can use pivot_longer from tidyr.
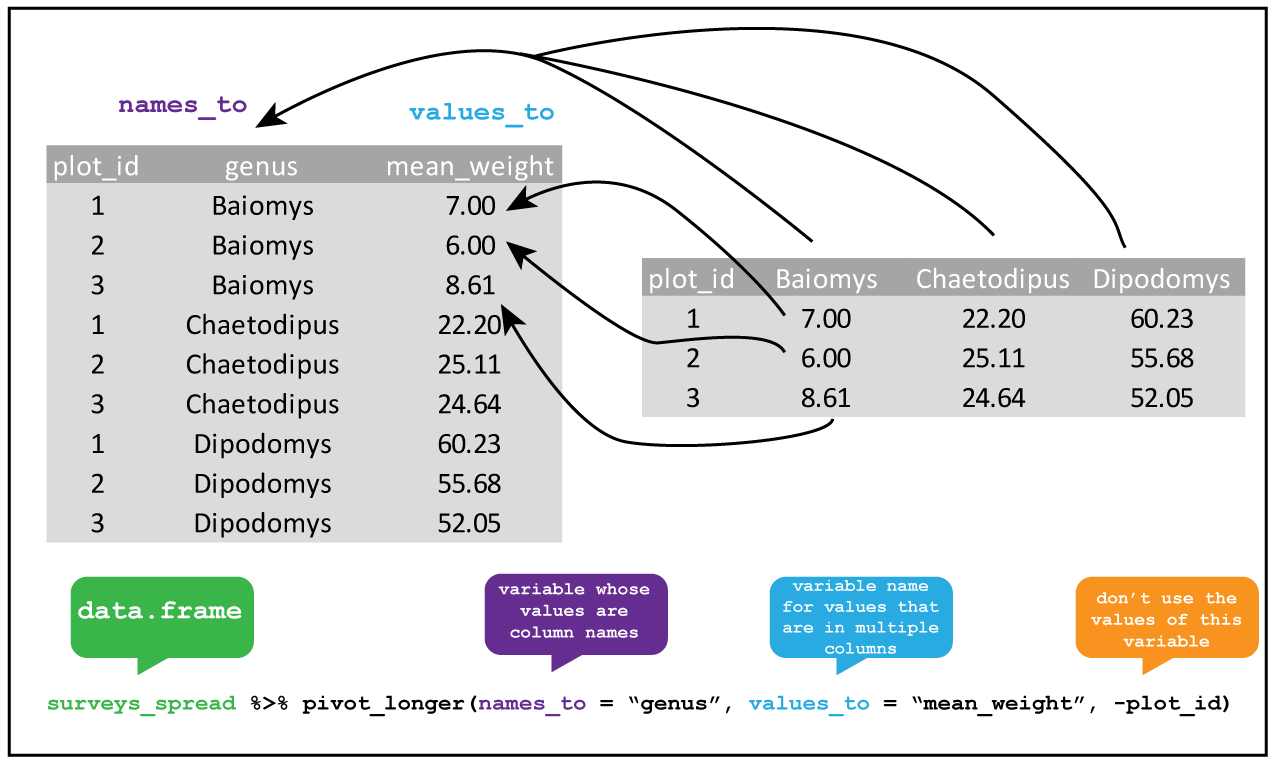
Figure 7.6: Pivoting longer from wide data.
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg)) %>%
pivot_longer(cols = 3:22, names_to = "metabolite_time", values_to = "abundance")## # A tibble: 180 × 4
## culture dose_mg metabolite_time abundance
## <chr> <dbl> <chr> <dbl>
## 1 e coli 0 glutamate_Time_0min 191666
## 2 e coli 0 glutamate_Time_10min 2082823
## 3 e coli 0 glutamate_Time_20min 1796910
## 4 e coli 0 glutamate_Time_50min 846402
## 5 e coli 0 glutamate_Time_2hr 1782879
## 6 e coli 0 lysine_Time_0min 1418
## 7 e coli 0 lysine_Time_10min 2004
## 8 e coli 0 lysine_Time_20min 1498
## 9 e coli 0 lysine_Time_50min 1356
## 10 e coli 0 lysine_Time_2hr 962
## # ℹ 170 more rowsAnother happy table…but STILL has multiple data encoded in a single column!
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg)) %>%
pivot_longer(cols = 3:22, names_to = "metabolite_time", values_to = "abundance") %>%
separate(metabolite_time, c("metabolite", "time_min"), sep="_")## # A tibble: 180 × 5
## culture dose_mg metabolite time_min abundance
## <chr> <dbl> <chr> <chr> <dbl>
## 1 e coli 0 glutamate Time 191666
## 2 e coli 0 glutamate Time 2082823
## 3 e coli 0 glutamate Time 1796910
## 4 e coli 0 glutamate Time 846402
## 5 e coli 0 glutamate Time 1782879
## 6 e coli 0 lysine Time 1418
## 7 e coli 0 lysine Time 2004
## 8 e coli 0 lysine Time 1498
## 9 e coli 0 lysine Time 1356
## 10 e coli 0 lysine Time 962
## # ℹ 170 more rowsAlmost there, but notice how metabolite_time has three items when we separate by _?
glutamate_Time_0min -> glutamateTime0min
We only provided two column names, though. Perhaps we can rethink our separator.
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg)) %>%
pivot_longer(cols = 3:22, names_to = "metabolite_time", values_to = "abundance") %>%
separate(metabolite_time, c("metabolite", "time_min"), sep="_Time_")## # A tibble: 180 × 5
## culture dose_mg metabolite time_min abundance
## <chr> <dbl> <chr> <chr> <dbl>
## 1 e coli 0 glutamate 0min 191666
## 2 e coli 0 glutamate 10min 2082823
## 3 e coli 0 glutamate 20min 1796910
## 4 e coli 0 glutamate 50min 846402
## 5 e coli 0 glutamate 2hr 1782879
## 6 e coli 0 lysine 0min 1418
## 7 e coli 0 lysine 10min 2004
## 8 e coli 0 lysine 20min 1498
## 9 e coli 0 lysine 50min 1356
## 10 e coli 0 lysine 2hr 962
## # ℹ 170 more rowsOk a new problem has appeared: mixed time units in our time_min column. We have both “min” and “hr”. We need to standardize to understand these as time units. We can mutate with case_when, which does specific things conditionally on particular observations in the tibble. This is a dplyr way of doing if statements. We will also advance it up by doing the “character” to “numeric” conversion in one go!
tbl_met_tidy <- tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg)) %>%
pivot_longer(cols = 3:22, names_to = "metabolite_time", values_to = "abundance") %>%
separate(metabolite_time, c("metabolite", "time_min"), sep="_Time_") %>%
mutate(
time_min = case_when(
str_detect(time_min, "min") ~ parse_number(time_min),
str_detect(time_min, "hr") ~ parse_number(time_min) * 60
)
)
tbl_met_tidy## # A tibble: 180 × 5
## culture dose_mg metabolite time_min abundance
## <chr> <dbl> <chr> <dbl> <dbl>
## 1 e coli 0 glutamate 0 191666
## 2 e coli 0 glutamate 10 2082823
## 3 e coli 0 glutamate 20 1796910
## 4 e coli 0 glutamate 50 846402
## 5 e coli 0 glutamate 120 1782879
## 6 e coli 0 lysine 0 1418
## 7 e coli 0 lysine 10 2004
## 8 e coli 0 lysine 20 1498
## 9 e coli 0 lysine 50 1356
## 10 e coli 0 lysine 120 962
## # ℹ 170 more rowsNow we can examine our tibble and confirm that once and for all, it is tidy. Let’s save it for later use. We can also pipe in our saving.
write_
tbl_met %>%
separate(Culture, c("culture", "dose_mg"), sep = " dose:") %>%
mutate(dose_mg = str_remove(dose_mg, "mg")) %>%
mutate(dose_mg = as.numeric(dose_mg)) %>%
pivot_longer(cols = 3:22, names_to = "metabolite_time", values_to = "abundance") %>%
separate(metabolite_time, c("metabolite", "time_min"), sep="_Time_") %>%
mutate(
time_min = case_when(
str_detect(time_min, "min") ~ parse_number(time_min),
str_detect(time_min, "hr") ~ parse_number(time_min) * 60
)
) %>%
write_csv("data/tidyed-met-df.csv")Non-pipe
One thing you can notice is how the dplyr verbs add up. We can continually pipe one data frame through a series of wrangling steps until we are satisfied. Operating in this way organizes processes in a way that is easy to manage. We could also do this:
# read the data
no_pipe_tbl_met <- read_csv("data/bacterial-metabolites_dose-simicillin_wide.csv")
no_pipe_tbl_met <- separate(no_pipe_tbl_met, Culture, c("culture", "dose_mg"), sep = " dose:")
no_pipe_tbl_met <- mutate(no_pipe_tbl_met, dose_mg = str_remove(dose_mg, "mg"))
no_pipe_tbl_met <- mutate(no_pipe_tbl_met, dose_mg = as.numeric(dose_mg))
no_pipe_tbl_met <- pivot_longer(no_pipe_tbl_met, cols = 3:22, names_to = "metabolite_time", values_to = "abundance")
no_pipe_tbl_met <- separate(no_pipe_tbl_met, metabolite_time, c("metabolite", "time_min"), sep="_Time_")
no_pipe_tbl_met_tidy <- mutate(no_pipe_tbl_met,
time_min = case_when(
str_detect(time_min, "min") ~ parse_number(time_min),
str_detect(time_min, "hr") ~ parse_number(time_min) * 60
)
)
no_pipe_tbl_met_tidy## # A tibble: 180 × 5
## culture dose_mg metabolite time_min abundance
## <chr> <dbl> <chr> <dbl> <dbl>
## 1 e coli 0 glutamate 0 191666
## 2 e coli 0 glutamate 10 2082823
## 3 e coli 0 glutamate 20 1796910
## 4 e coli 0 glutamate 50 846402
## 5 e coli 0 glutamate 120 1782879
## 6 e coli 0 lysine 0 1418
## 7 e coli 0 lysine 10 2004
## 8 e coli 0 lysine 20 1498
## 9 e coli 0 lysine 50 1356
## 10 e coli 0 lysine 120 962
## # ℹ 170 more rowssummarize
Now we have a tidy tibble, we can summarize very rapidly.
## # A tibble: 12 × 3
## # Groups: culture [3]
## culture metabolite mean
## <chr> <chr> <dbl>
## 1 e coli glutamate 2323045.
## 2 e coli lysine 1519827.
## 3 e coli phosphatidylcholine 67749754.
## 4 e coli succinic acid 1906850.
## 5 p aeruginosa glutamate 2902178
## 6 p aeruginosa lysine 2336119.
## 7 p aeruginosa phosphatidylcholine 77078132.
## 8 p aeruginosa succinic acid 3525432.
## 9 staph aureus glutamate 226736.
## 10 staph aureus lysine 193944.
## 11 staph aureus phosphatidylcholine 5836359.
## 12 staph aureus succinic acid 133337.What other questions can we ask???
Exercises
 |
|
- Download the data.
url <- "https://raw.githubusercontent.com/jeffsocal/ASMS_R_Basics/main/data/bacterial-metabolites_dose-simicillin_messy.csv"
download.file(url, destfile = "./data/bacterial-metabolites_dose-simicillin_messy.csv")- Read in the messy bacteria data and store it as a variable.
## # A tibble: 9 × 12
## Culture User glutamate_runtime_0hr glutamate_runtime_24hr
## <chr> <chr> <dbl> <dbl>
## 1 e coli dose--0-mg/ml Mass… 191666 2082823
## 2 staph aureus dose--0-mg/… Mass… 125210 132156
## 3 p aeruginosa dose--0-mg/… Mass… 978126 397012
## 4 e coli dose--10-mg/ml Mass… 1445 1017467
## 5 staph aureus dose--10-mg… Mass… 152 128683
## 6 p aeruginosa dose--10-mg… Mass… 23 1612622
## 7 e coli dose--20-mg/ml Mass… 885 42356
## 8 staph aureus dose--20-mg… Mass… 136 4276
## 9 p aeruginosa dose--20-mg… Mass… 3098 68146
## # ℹ 8 more variables: glutamate_runtime_2day <dbl>,
## # glutamate_runtime_3day <dbl>, glutamate_runtime_5day <dbl>,
## # lysine_runtime_0hr <dbl>, lysine_runtime_24hr <dbl>,
## # lysine_runtime_2day <dbl>, lysine_runtime_3day <dbl>,
## # lysine_runtime_5day <dbl>In all proceeding exercises, pipe results from previous exercise into current exercise creating a single lone pipe for data processing
- Separate
Culturecolumn containing culture and dose intocultureanddose_mg_mlcolumns.
## # A tibble: 9 × 13
## culture dose_mg_ml User glutamate_runtime_0hr glutamate_runtime_24hr
## <chr> <chr> <chr> <dbl> <dbl>
## 1 e coli 0-mg/ml Mass S… 191666 2082823
## 2 staph aureus 0-mg/ml Mass S… 125210 132156
## 3 p aeruginosa 0-mg/ml Mass S… 978126 397012
## 4 e coli 10-mg/ml Mass S… 1445 1017467
## 5 staph aureus 10-mg/ml Mass S… 152 128683
## 6 p aeruginosa 10-mg/ml Mass S… 23 1612622
## 7 e coli 20-mg/ml Mass S… 885 42356
## 8 staph aureus 20-mg/ml Mass S… 136 4276
## 9 p aeruginosa 20-mg/ml Mass S… 3098 68146
## # ℹ 8 more variables: glutamate_runtime_2day <dbl>,
## # glutamate_runtime_3day <dbl>, glutamate_runtime_5day <dbl>,
## # lysine_runtime_0hr <dbl>, lysine_runtime_24hr <dbl>,
## # lysine_runtime_2day <dbl>, lysine_runtime_3day <dbl>,
## # lysine_runtime_5day <dbl>- Make
dose_mg_mlcolumn numeric by removing the text and change the column data type from character to numeric.
## # A tibble: 9 × 13
## culture dose_mg_ml User glutamate_runtime_0hr glutamate_runtime_24hr
## <chr> <dbl> <chr> <dbl> <dbl>
## 1 e coli 0 Mass S… 191666 2082823
## 2 staph aureus 0 Mass S… 125210 132156
## 3 p aeruginosa 0 Mass S… 978126 397012
## 4 e coli 10 Mass S… 1445 1017467
## 5 staph aureus 10 Mass S… 152 128683
## 6 p aeruginosa 10 Mass S… 23 1612622
## 7 e coli 20 Mass S… 885 42356
## 8 staph aureus 20 Mass S… 136 4276
## 9 p aeruginosa 20 Mass S… 3098 68146
## # ℹ 8 more variables: glutamate_runtime_2day <dbl>,
## # glutamate_runtime_3day <dbl>, glutamate_runtime_5day <dbl>,
## # lysine_runtime_0hr <dbl>, lysine_runtime_24hr <dbl>,
## # lysine_runtime_2day <dbl>, lysine_runtime_3day <dbl>,
## # lysine_runtime_5day <dbl>- Pivot the table from wide to long creating
metabolite,time_hr&abundancecolumns.
## # A tibble: 90 × 6
## culture dose_mg_ml User metabolite time_hr abundance
## <chr> <dbl> <chr> <chr> <chr> <dbl>
## 1 e coli 0 Mass Spectrometrist glutamate 0hr 191666
## 2 e coli 0 Mass Spectrometrist glutamate 24hr 2082823
## 3 e coli 0 Mass Spectrometrist glutamate 2day 1796910
## 4 e coli 0 Mass Spectrometrist glutamate 3day 846402
## 5 e coli 0 Mass Spectrometrist glutamate 5day 1782879
## 6 e coli 0 Mass Spectrometrist lysine 0hr 1418
## 7 e coli 0 Mass Spectrometrist lysine 24hr 2004
## 8 e coli 0 Mass Spectrometrist lysine 2day 1498
## 9 e coli 0 Mass Spectrometrist lysine 3day 1356
## 10 e coli 0 Mass Spectrometrist lysine 5day 961
## # ℹ 80 more rows- Make sure
time_hrcontains just hours and not a mixture of days and hours.
## # A tibble: 90 × 6
## culture dose_mg_ml User metabolite time_hr abundance
## <chr> <dbl> <chr> <chr> <dbl> <dbl>
## 1 e coli 0 Mass Spectrometrist glutamate 0 191666
## 2 e coli 0 Mass Spectrometrist glutamate 24 2082823
## 3 e coli 0 Mass Spectrometrist glutamate 48 1796910
## 4 e coli 0 Mass Spectrometrist glutamate 72 846402
## 5 e coli 0 Mass Spectrometrist glutamate 120 1782879
## 6 e coli 0 Mass Spectrometrist lysine 0 1418
## 7 e coli 0 Mass Spectrometrist lysine 24 2004
## 8 e coli 0 Mass Spectrometrist lysine 48 1498
## 9 e coli 0 Mass Spectrometrist lysine 72 1356
## 10 e coli 0 Mass Spectrometrist lysine 120 961
## # ℹ 80 more rows- Remove the
Usercolumn. See cheatsheet here: data-wrangling-cheatsheet or consult internet.
## # A tibble: 90 × 5
## culture dose_mg_ml metabolite time_hr abundance
## <chr> <dbl> <chr> <dbl> <dbl>
## 1 e coli 0 glutamate 0 191666
## 2 e coli 0 glutamate 24 2082823
## 3 e coli 0 glutamate 48 1796910
## 4 e coli 0 glutamate 72 846402
## 5 e coli 0 glutamate 120 1782879
## 6 e coli 0 lysine 0 1418
## 7 e coli 0 lysine 24 2004
## 8 e coli 0 lysine 48 1498
## 9 e coli 0 lysine 72 1356
## 10 e coli 0 lysine 120 961
## # ℹ 80 more rows